Import the records once the download is finished; this could take a while on a slow connection.
tic () # starts a timer <- occ_download_get ('0066939-241126133413365' ) # generated in `Fetching plant occurrence records from GBIF`
Download file size: 1934.4 MB
file exists & overwrite=FALSE, not overwriting...
<- occ_download_import (records, select= c ("scientificName" ,"species" ,"taxonKey" , "speciesKey" ,"year" ,"decimalLongitude" ,"decimalLatitude" ,"countryCode" ))toc () # ends the timer
Inspect the lat/long values.
cat ("Longitude extremes:" , min (data$ decimalLongitude), max (data$ decimalLongitude))
Longitude extremes: -179.9753 179.9876
cat ("Latitude extremes:" , min (data$ decimalLatitude), max (data$ decimalLatitude))
Latitude extremes: -55.04704 85.75
Filter out points at the edges of flat map, they are causes some stretching issues when building hex bins.
<- data %>% filter (between (decimalLatitude, - 85 , 85 )) %>% filter (between (decimalLongitude, - 175 , 175 ))
Convert 18.5 Mil records into H3 hex shapes
We use the Uber H3 hexagon system for binning records into spatial areas to plot densities.
Convert df to sf
<- st_as_sf (x = data2, coords = c ("decimalLongitude" , "decimalLatitude" ),crs = 4326 )
Check # of species vs # of scientific name
cat (length (unique (data_sf$ scientificName)), length (unique (data_sf$ species)), length (unique (data_sf$ taxonKey)), length (unique (data_sf$ speciesKey)))
<- as.data.frame (unique (data_sf$ species))
unique(data_sf$species)
1 Holcus lanatus
2 Senecio vulgaris
3 Hypochaeris radicata
4 Achillea millefolium
5 Agrostis stolonifera
6 Juncus articulatus
7 Prunella vulgaris
8 Chamaenerion angustifolium
9 Deschampsia cespitosa
10 Spergularia rubra
11 Trifolium repens
12 Ranunculus acris
13 Ranunculus sceleratus
14 Eleocharis acicularis
15 Festuca rubra
16 Phalaris arundinacea
17 Phleum pratense
18 Sonchus asper
19 Eleocharis palustris
20 Poa pratensis
21 Rubus idaeus
22 Sonchus arvensis
23 Potamogeton gramineus
24 Cerastium arvense
25 Carex canescens
26 Galium boreale
27 Juniperus communis
28 Tragopogon dubius
29 Bromus tectorum
30 Erigeron acris
31 Thlaspi arvense
32 Campanula rotundifolia
33 Onopordum acanthium
34 Limosella aquatica
35 Asparagus officinalis
36 Veronica peregrina
37 Koeleria macrantha
38 Poa palustris
39 Dianthus armeria
40 Solidago canadensis
41 Alyssum alyssoides
42 Lepidium virginicum
43 Myosurus minimus
44 Allium schoenoprasum
45 Euphorbia esula
46 Chondrilla juncea
47 Bromus inermis
48 Potentilla norvegica
49 Xanthium strumarium
50 Agrostis scabra
51 Carex limosa
52 Poa alpina
53 Phleum alpinum
54 Astragalus alpinus
55 Myosotis alpestris
56 Eritrichium nanum
57 Potentilla argentea
58 Alyssum simplex
59 Oxyria digyna
60 Sagina saginoides
61 Anthoxanthum nitens
62 Bromus squarrosus
63 Dasiphora fruticosa
64 Persicaria amphibia
65 Viola adunca
66 Collinsia parviflora
67 Agrostis exarata
68 Lepidium appelianum
69 Erythranthe lewisii
70 Ranunculus eschscholtzii
71 Hesperochiron pumilus
72 Maianthemum racemosum
73 Actaea rubra
74 Lithophragma parviflorum
75 Acer glabrum
76 Aconitum columbianum
77 Taraxacum officinale
78 Populus tremuloides
79 Fragaria virginiana
80 Iris missouriensis
81 Aquilegia flavescens
82 Sedum lanceolatum
83 Mahonia repens
84 Polemonium pulcherrimum
85 Ceratocephala orthoceras
86 Spiranthes romanzoffiana
87 Ranunculus uncinatus
88 Veronica americana
89 Allium cernuum
90 Platanthera dilatata
91 Ranunculus alismifolius
92 Elymus glaucus
93 Viola nuttallii
94 Neottia cordata
95 Wyethia sagittata
96 Linum lewisii
97 Geranium viscosissimum
98 Carex duriuscula
99 Agropyron cristatum
100 Phlox longifolia
101 Lysimachia ciliata
102 Bromus japonicus
103 Sporobolus cryptandrus
104 Festuca octoflora
105 Elymus smithii
106 Danthonia spicata
107 Sphaeralcea coccinea
108 Equisetum laevigatum
109 Tamarix chinensis
110 Bromus carinatus
111 Elymus elymoides
112 Hesperostipa comata
113 Plantago elongata
114 Polygonum ramosissimum
115 Castilleja linariifolia
116 Glyceria borealis
117 Calamagrostis canadensis
118 Geranium richardsonii
119 Pseudoroegneria spicata
120 Schoenoplectus acutus
121 Boechera stricta
122 Camelina microcarpa
123 Hordeum jubatum
124 Gayophytum diffusum
125 Potentilla hippiana
126 Polygonum douglasii
127 Eleocharis rostellata
128 Torreyochloa pallida
129 Collomia linearis
130 Elymus violaceus
131 Pinus contorta
132 Rumex fueginus
133 Schoenoplectus subterminalis
134 Androsace occidentalis
135 Descurainia incana
136 Delphinium nuttallianum
137 Eriogonum umbellatum
138 Androsace septentrionalis
139 Carex obtusata
140 Senecio triangularis
141 Beckmannia syzigachne
142 Taraxacum erythrospermum
143 Poa glauca
144 Koeleria spicata
145 Taraxacum ceratophorum
146 Phlox gracilis
147 Scolochloa festucacea
148 Salvia nemorosa
149 Anemone multifida
150 Deschampsia danthonioides
151 Poa secunda
152 Penstemon fruticosus
153 Packera cana
154 Potentilla gordonii
155 Carex vallicola
156 Veronica wormskjoldii
157 Delphinium occidentale
158 Symphyotrichum ascendens
159 Lupinus argenteus
160 Leptosiphon septentrionalis
161 Puccinellia nuttalliana
162 Symphyotrichum bracteolatum
163 Erigeron caespitosus
164 Helianthus nuttallii
165 Carex pachystachya
166 Symphyotrichum foliaceum
167 Geum triflorum
168 Oxytropis sericea
169 Carex simulata
170 Mertensia ciliata
171 Festuca brachyphylla
172 Eremogone congesta
173 Penstemon deustus
174 Rumex paucifolius
175 Helianthella uniflora
176 Astragalus miser
177 Eriogonum heracleoides
178 Tetradymia canescens
179 Carex rossii
180 Carex brunnescens
181 Erigeron glabellus
182 Calochortus eurycarpus
183 Astragalus agrestis
184 Carex petasata
185 Poa cusickii
186 Lomatium triternatum
187 Chenopodiastrum simplex
188 Polemonium occidentale
189 Castilleja rhexifolia
190 Carex disperma
191 Erigeron pumilus
192 Antennaria microphylla
193 Agoseris glauca
194 Scrophularia lanceolata
195 Castilleja miniata
196 Plagiobothrys scouleri
197 Astragalus purshii
198 Taraxia subacaulis
199 Stenotus acaulis
200 Eremopyrum triticeum
201 Boechera retrofracta
202 Leymus cinereus
203 Micranthes rhomboidea
204 Phlox multiflora
205 Eriocoma lettermanii
206 Alyssum turkestanicum
207 Cherleria obtusiloba
208 Senecio crassulus
209 Carex chalciolepis
210 Carex elynoides
211 Clematis hirsutissima
212 Heterotheca villosa
213 Poa arida
214 Draba albertina
215 Polygonum polygaloides
216 Lomatium macrocarpum
217 Iliamna rivularis
218 Carex praegracilis
219 Draba nemorosa
220 Danthonia intermedia
221 Penstemon attenuatus
222 Castilleja pilosa
223 Festuca idahoensis
224 Penstemon procerus
225 Veronica biloba
226 Epilobium hornemannii
227 Myosurus apetalus
228 Draba oligosperma
229 Poa interior
230 Crepis modocensis
231 Orthocarpus luteus
232 Drymocallis pseudorupestris
233 Toxicoscordion venenosum
234 Eriogonum flavum
235 Carex raynoldsii
236 Erigeron ochroleucus
237 Draba crassifolia
238 Penstemon cyaneus
239 Elymus scribneri
240 Carex spectabilis
241 Chenopodium atrovirens
242 Saxifraga bronchialis
243 Solidago multiradiata
244 Poa reflexa
245 Eriogonum caespitosum
246 Stephanomeria runcinata
247 Carex filifolia
248 Geum macrophyllum
249 Muhlenbergia richardsonis
250 Festuca kingii
251 Castilleja pulchella
252 Calamagrostis purpurascens
253 Erigeron corymbosus
254 Thalictrum sparsiflorum
255 Thalictrum occidentale
256 Epilobium lactiflorum
257 Glyceria elata
258 Carex parryana
259 Oreocarya spiculifera
260 Fritillaria atropurpurea
261 Lactuca biennis
262 Allium brevistylum
263 Boechera lemmonii
264 Muhlenbergia glomerata
265 Draba globosa
266 Poa leptocoma
267 Wyethia helianthoides
268 Carex haydeniana
269 Carex paysonis
270 Claytonia multiscapa
271 Opuntia fragilis
272 Carex leporinella
273 Ionactis alpina
274 Eriocoma nelsonii
275 Symphyotrichum campestre
276 Carex hoodii
277 Taraxia breviflora
278 Carex neurophora
279 Arabis pycnocarpa
280 Ceanothus velutinus
281 Parnassia fimbriata
282 Silene parryi
283 Heuchera cylindrica
284 Dryas octopetala
285 Scheuchzeria palustris
286 Synthyris wyomingensis
287 Telesonix heucheriformis
288 Heuchera parviflora
289 Draba cana
290 Calamagrostis stricta
291 Polygonum achoreum
292 Draba incerta
293 Carex tahoensis
294 Ventenata dubia
295 Pedicularis cystopteridifolia
296 Castilleja crista-galli
297 Conimitella williamsii
298 Bromus porteri
299 Hieracium praealtum
300 Filago arvensis
301 Matricaria discoidea
302 Blitum nuttallianum
303 Koeleria vaseyi
304 Draba praealta
305 Arabis nuttallii
306 Mimulus moschatus
307 Physaria reediana
308 Eriocoma richardsonii
309 Carex stenoptila
310 Ranunculus mongolicus
311 Castilleja nivea
Get CRS info.
[1] "GEOGCRS[\"WGS 84\",\n ENSEMBLE[\"World Geodetic System 1984 ensemble\",\n MEMBER[\"World Geodetic System 1984 (Transit)\"],\n MEMBER[\"World Geodetic System 1984 (G730)\"],\n MEMBER[\"World Geodetic System 1984 (G873)\"],\n MEMBER[\"World Geodetic System 1984 (G1150)\"],\n MEMBER[\"World Geodetic System 1984 (G1674)\"],\n MEMBER[\"World Geodetic System 1984 (G1762)\"],\n MEMBER[\"World Geodetic System 1984 (G2139)\"],\n ELLIPSOID[\"WGS 84\",6378137,298.257223563,\n LENGTHUNIT[\"metre\",1]],\n ENSEMBLEACCURACY[2.0]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n CS[ellipsoidal,2],\n AXIS[\"geodetic latitude (Lat)\",north,\n ORDER[1],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n AXIS[\"geodetic longitude (Lon)\",east,\n ORDER[2],\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n USAGE[\n SCOPE[\"Horizontal component of 3D system.\"],\n AREA[\"World.\"],\n BBOX[-90,-180,90,180]],\n ID[\"EPSG\",4326]]"
This step pairs lat long into a point geometry.
st_transform (data_sf, 4326 )
Simple feature collection with 20503896 features and 6 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -174.9518 ymin: -55.04704 xmax: 175 ymax: 84.75
Geodetic CRS: WGS 84
# A tibble: 20,503,896 × 7
scientificName species taxonKey speciesKey year countryCode
* <chr> <chr> <int> <int> <int> <chr>
1 Holcus lanatus L. Holcus lanatus 2706164 2706164 2018 FR
2 Holcus lanatus L. Holcus lanatus 2706164 2706164 2018 FR
3 Senecio vulgaris L. Senecio vulgar… 3108983 3108983 1985 FR
4 Hypochaeris radicata L. Hypochaeris ra… 3093702 3093702 2019 FR
5 Achillea millefolium L. Achillea mille… 3120060 3120060 2015 FR
6 Agrostis stolonifera L. Agrostis stolo… 2706435 2706435 1994 FR
7 Achillea millefolium L. Achillea mille… 3120060 3120060 2000 FR
8 Senecio vulgaris L. Senecio vulgar… 3108983 3108983 2009 FR
9 Juncus articulatus L. Juncus articul… 2701261 2701261 1991 FR
10 Prunella vulgaris L. Prunella vulga… 5341297 5341297 1999 FR
# ℹ 20,503,886 more rows
# ℹ 1 more variable: geometry <POINT [°]>
Toss records without a species assigned and randomly subsample per species.
<- data_sf %>% filter (species != "" ) %>% group_by (species) %>% #slice_sample(n = params$random_sample_n) # number of samples slice_sample (prop = params$ random_sample_prop) # proportion of samples
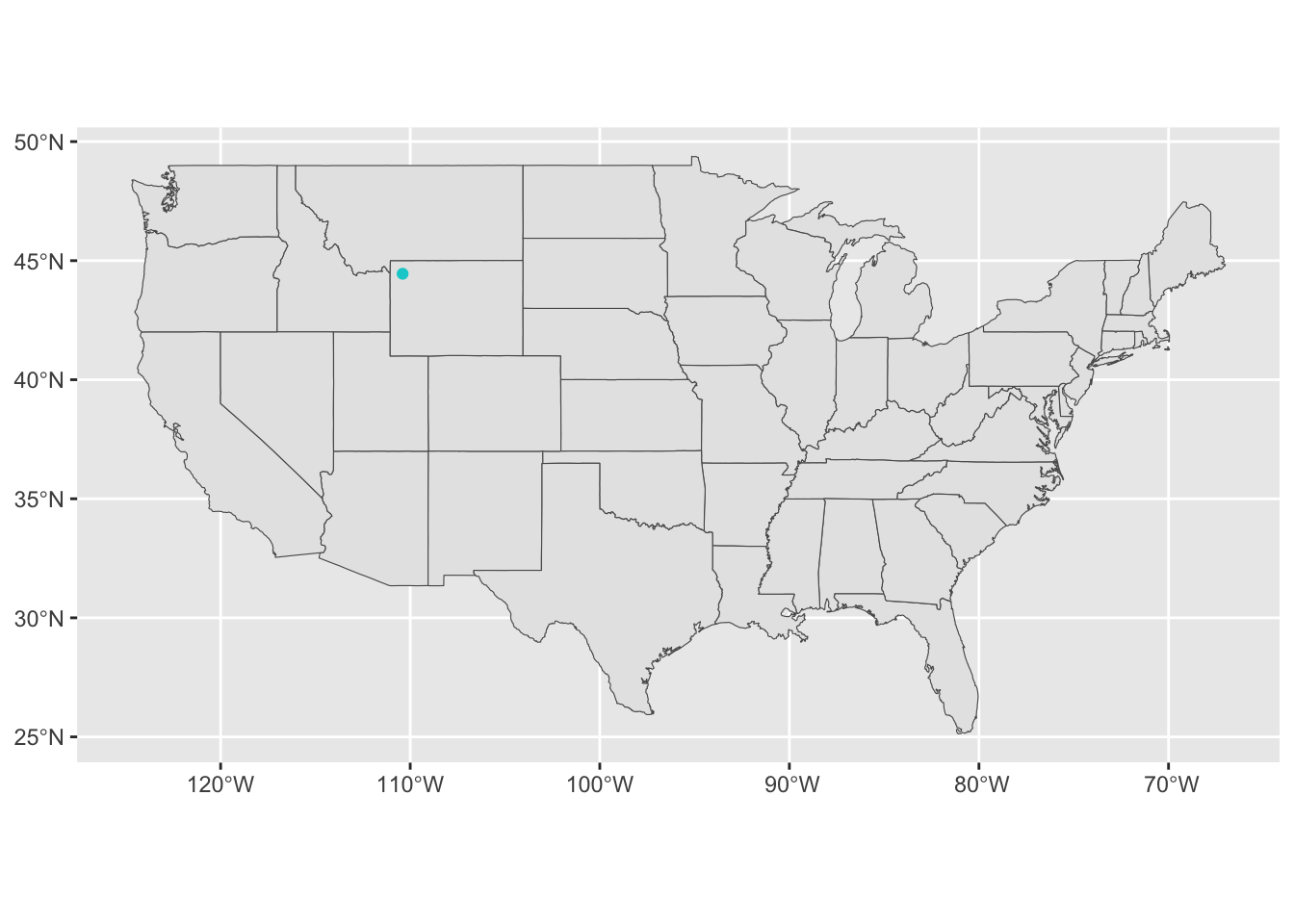
Make a point for the centroid of YNP (middle of Yellowstone Lake), to use for distance calculations.
<- st_point (c (- 110.40 , 44.45 )) %>% st_coordinates () %>% as.data.frame () %>% st_as_sf (coords = c ("X" , "Y" ), crs = 4326 )= st_as_sf (map ('state' , plot = FALSE , fill = TRUE ))ggplot () + geom_sf (data = usa) + geom_sf (data = YNP_centroid, aes (geometry= geometry), pch = 19 , color = 'darkturquoise' )
This block will calculate the distances from the center of Yellowstone for each row in the dataset.
tic ()<- data_sf %>% mutate (dist = st_distance (geometry, YNP_centroid) %>% set_units ("km" ))beep (2 )toc ()
Summarize the distances to get a count, average distance, and sd of distance for each species.
This will output a dataframe where each row in one of the 279 species with occurrence records in GBIF. The geometries are also merged to a multipoint so a minimum convex polygon for a single species could be plotted easily.
tic ()<- data_sf %>% group_by (species) %>% summarise (count = n (),min_dist = min (dist),max_dist = max (dist),mean_dist = mean (dist),sd_dist = sd (dist)beep (2 )summary (species_dist)
species count min_dist max_dist
Length:311 Min. : 26 Min. : 1.268 Min. : 174
Class :character 1st Qu.: 1083 1st Qu.: 12.825 1st Qu.: 3099
Mode :character Median : 3751 Median : 31.953 Median : 8666
Mean : 65929 Mean : 61.950 Mean : 8562
3rd Qu.: 15874 3rd Qu.: 53.920 3rd Qu.:12698
Max. :1392917 Max. :5688.516 Max. :19489
mean_dist sd_dist geometry
Min. : 108.5 Min. : 30.38 MULTIPOINT :311
1st Qu.: 710.5 1st Qu.: 328.71 epsg:4326 : 0
Median :1068.3 Median : 786.85 +proj=long...: 0
Mean :2889.7 Mean :1124.93
3rd Qu.:6019.3 3rd Qu.:1576.01
Max. :8737.6 Max. :5122.90
Output a file with summary data about distances!
%>% st_drop_geometry () %>% write.csv ("./distance_from_YNP_by_species.csv" )
Use this to get a few CRS codes to try for projection if things look off.
# A tibble: 10 × 6
crs_code crs_name crs_type crs_gcs crs_units crs_proj4
<chr> <chr> <chr> <dbl> <chr> <chr>
1 6931 WGS 84 / NSIDC EASE-Grid 2.0 N… project… 4326 m +proj=la…
2 3395 WGS 84 / World Mercator project… 4326 m +proj=me…
3 3857 WGS 84 / Pseudo-Mercator project… 4326 m +proj=me…
4 6933 WGS 84 / NSIDC EASE-Grid 2.0 G… project… 4326 m +proj=ce…
5 3832 WGS 84 / PDC Mercator project… 4326 m +proj=me…
6 6932 WGS 84 / NSIDC EASE-Grid 2.0 S… project… 4326 m +proj=la…
7 8903 RGWF96 / UTM zone 1S project… 8900 m +proj=ut…
8 3576 WGS 84 / North Pole LAEA Russia project… 4326 m +proj=la…
9 3575 WGS 84 / North Pole LAEA Europe project… 4326 m +proj=la…
10 3574 WGS 84 / North Pole LAEA Atlan… project… 4326 m +proj=la…
Pull geospatial hex locations for each point set.
$ h3_index <- geo_to_h3 (data_sf, params$ resolution)
Group by hex bin and count species in each one.
# Check that the number of unique bins is much less than the number of rows length (unique (data_sf$ h3_index))<- data_sf %>% group_by (h3_index) %>% summarise (distinct_species = n_distinct (species))beep (2 )
Count the number of records in each hex bin.
<- as.data.frame (table (data_sf2$ h3_index))
Get the coordinates of of each hex bin.
<- h3_to_geo_boundary_sf (data_sf2$ h3_index)
Merge the bin coords with frequency data for plotting.
<- merge (as.data.frame (data_sf2), data_hex, by= "h3_index" )
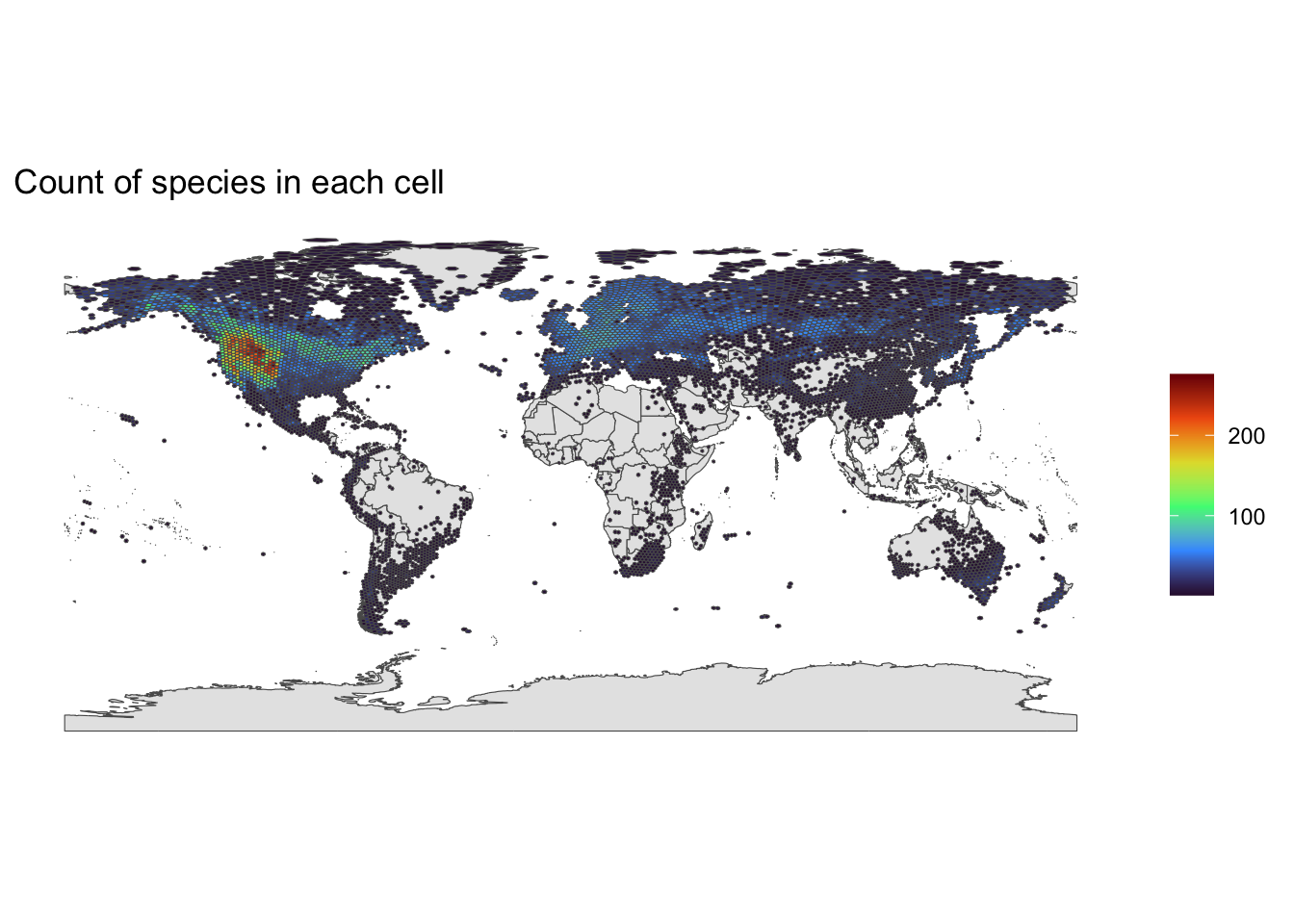
Report out the number of hexagons, and the min and max number of species in any one hex.
cat (length (unique (plot_data$ h3_index)), min (plot_data$ distinct_species), max (plot_data$ distinct_species))
Build the map
<- ne_countries (scale = 'large' , returnclass = 'sf' )
ggplot () + geom_sf (data = worldmap$ geometry) + geom_sf (data = plot_data, aes (geometry = geometry.y, fill= distinct_species)) + scale_fill_viridis_c (option= "turbo" ) + ggtitle ("Count of species in each cell" ) + theme_classic () + theme (legend.title = element_blank ()) ggsave ("BOLD_GBIF_12400km2.png" ) # uncomment to save a copy